Co-localization analyses of genomic features: Methods and potential pitfalls
Chakravarthi Kanduri(skanduri@ifi.uio.no), Christoph Bock(cbock@cemm.oeaw.ac.at), Sveinung Gundersen (sveinungu@ifi.uio.no), Eivind Hovig (ehovig@ifi.uio.no), Geir Kjetil Sandve(geirksa@ifi.uio.no)
(This page provides examples of the pitfalls related to co-localization analysis of genomic features. All the examples are provided through the Genomic HyperBrowser, which is tightly connected to the Galaxy framework. Users not familiar with Galaxy framework can quickly get familiar by following a quick introduction tutorial here (https://galaxyproject.org/tutorials/g101/).)
Example-9: Consider the possibility of co-localizing confounding features
The spatial and functional dependencies that exist between various genomic elements is a known phenomenon (see main article). In some cases, the statistical association of two functional genomic elements may, in fact, be driven by their co-localization with a third genomic element that has not been tested for. Not testing for the potential confounding factors in certain cases may lead to incomplete conclusions.
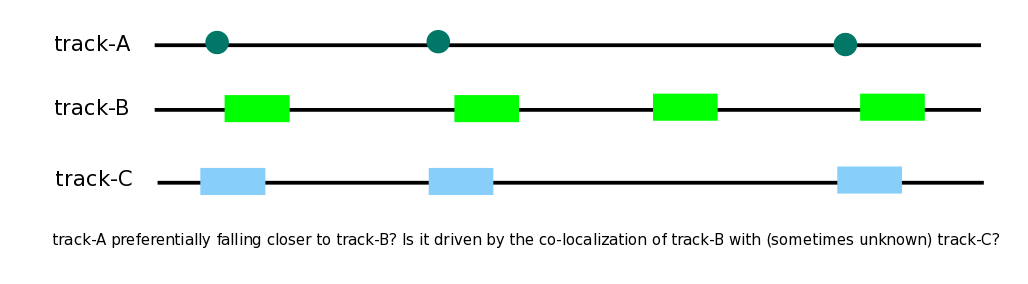
Here we demonstrate this issue with an example. In the galaxy history below, history elements 1-3 contain the DNAse hotspots, H3K4ME1 sites, and TFBS sites of p300 from K562 cell line (ENCODE). There was a very strong evidence for the co-localization of p300 sites with both DNAse hotspots and H3K4ME1 enhancer activation marks (history elements 4-5). However, it is noteworthy that DNAse hypersensitive sites are known to strongly co-localize with enhancer activation marks in general, which is also demonstrated here in our analysis through an enrichment of the test statistic by ~ 7-fold (history element 6).
Author
hb-superuser
Related Pages
All published pages
Published pages by hb-superuser
Rating
(0 ratings, 0.0 average)
Tags