Co-localization analyses of genomic features: Methods and potential pitfalls
Chakravarthi Kanduri(skanduri@ifi.uio.no), Christoph Bock(cbock@cemm.oeaw.ac.at), Sveinung Gundersen (sveinungu@ifi.uio.no), Eivind Hovig (ehovig@ifi.uio.no), Geir Kjetil Sandve(geirksa@ifi.uio.no)
(This page provides examples of the pitfalls related to co-localization analysis of genomic features. All the examples are provided through the Genomic HyperBrowser, which is tightly connected to the Galaxy framework. Users not familiar with Galaxy framework can quickly get familiar by following a quick introduction tutorial here (https://galaxyproject.org/tutorials/g101/).)
Example-6: Be aware of the fact that all statistical tests assume a null model, and the conclusions of co-localization analysis vary depending on the choice of null model
Here, we demonstrate that the conclusions of co-localization analysis would vary depending upon the choice of null models. For this, we provide examples based on Monte Carlo simulations.
Monte Carlo (MC) -based approach of hypothesis testing relies on sampling a test statistic using permutation models. The null models can be defined in various ways in terms of the constraints imposed on the shuffling space. The simplest null model assumes uniformity and independence of the genomic locations being shuffled, while in reality genomic locations and events at individual base pairs are not independent and often found in clumps. The text in the null models section of the article and a separate case-study (reference 32 in main article) discussed the importance of assessing the consistency of observations with multiple null model choices. Also, it has been discussed that a conservative null model that retains the essential biological properties of the observed data would be a safer choice, when interpreting the findings.
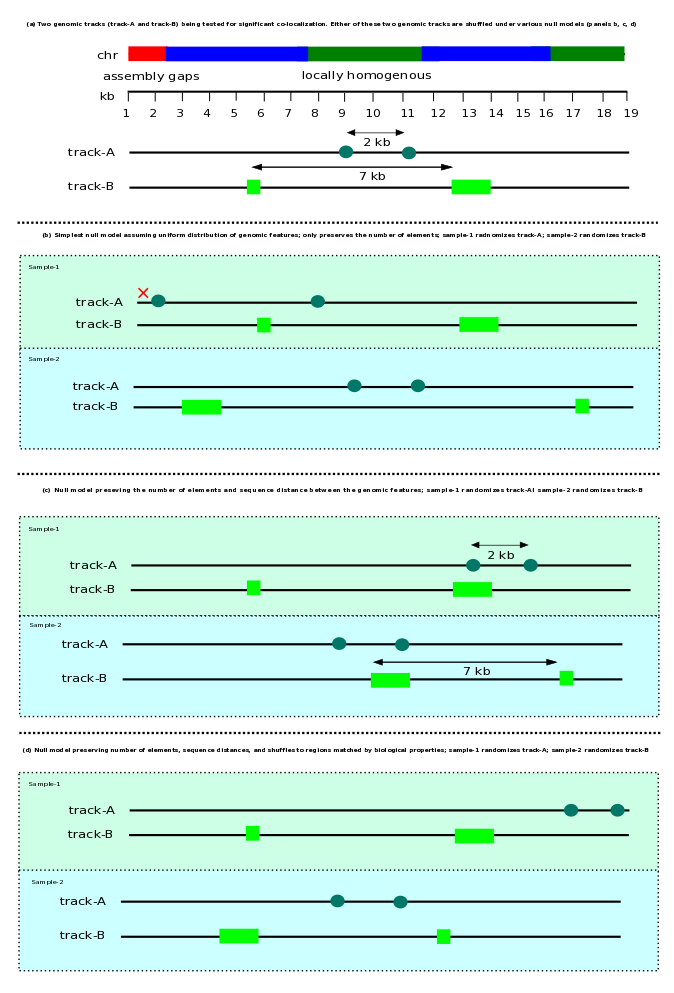
Here, as an example, we show the effect of different null models on the conclusions when studying the relationship between a set of GWAS-implicated SNPs and an enhancer activating mark. In the galaxy history below, history elements 1-9 involve downloading and processing of an enhancer activating mark H3K4ME1 from brain tissue (Roadmap Epigenomics). History elements 10 and 11 contain the final tracks used in analyses: H3K4ME1-brain track, and Schizophrenia GWAS SNPs track. History elements 14-18, using different definitions of null models, ask a question whether Schizophrenia-implicated SNPs fall inside H3K4ME1 mark more than expected by chance. Except for the simplest of the null model (history element 14), larger p-values were observed with an increase in the preservation of geometric properties (distance between the points or segments). When the observations across all the null models are considered (including the effect-size), one may conclude that there is no strong evidence against the null hypothesis.
Author
hb-superuser
Related Pages
All published pages
Published pages by hb-superuser
Rating
(0 ratings, 0.0 average)
Tags